Research | Overview
Gene regulation in normal hematopoiesis
The hematopoietic system serves a valuable model to study basic gene regulatory mechanisms involved in cell fate decisions and cell maturation. An adult human produces nearly a trillion new blood cells every day. This remarkable process is achieved by the self-renewal of pluripotent hematopoietic stem cells (HSCs) and their occasional entry into pathways of gradually restricted lineage potential. This eventually leads to the generation of more than 8 distinct functional cell types. Highly orchestrated gene expression changes occur during this differentiation process.
This involves the activation of lineage-specific gene programs and the simultaneous repression of stem/progenitor and alternate lineage gene programs. Abundant resources are available to study gene regulation in the hematopoietic system. These include inducible cell lines, easy access to primary cells, well-established in vitro differentiation protocols, mouse models, cell surface marker data and antibodies to separate hematopoietic cell populations, well-annotated gene expression data, transcription factor ChIP-seq datasets, chromatin accessibility maps, histone modification data, and other ENCODE data. Current research focus areas on hematopoietic gene regulation in the Cantor Lab include:
- chromatin interaction dynamics during hematopoiesis
- interplay of lineage-specific transcription factors, cohesin, CTCF and DNA methylation modifying enzymes to modulate 3D-chromatin structure
- role of cell signaling pathways in modulating transcription factor activity and chromatin occupancy
- locus-specific regulation of transcription factor activity via recruitment of different chromatin remodeling complexes.
- post-transcriptional control of lineage-specific transcription factors in hematopoiesis.
Huang H, Woo AJ, Waldon Z, Schindler Y, Moran TB, Zhu HH, Feng G-S, Steen H, Cantor AB. 2012. A Src Family Kinase-Shp2 Axis Controls RUNX1 Activity in Megakaryocyte and T Lymphocyte Differentiation. Genes Dev 26: 1587-1601;
Yu M, Mazor T, Huang H, Huang H-T, Kathrein KL, Woo AJ, Chouinard CR, Labadorf A, Akie TE, Moran TB, Xie H, Zacharek S, Taniuchi I, Roeder RG, Kim CF, Zon LI, Fraenkel E, Cantor AB. 2012. Direct Recruitment of Polycomb Repressive Complex 1 to Chromatin by Core Binding Transcription Factors. Molecular Cell 45:330-343. [NIHMS 349597].
Yu M, Riva L, Xie H, Schindler Y, Moran TB, Cheng Y, Yu D, Hardison R, Weiss MJ, Orkin SH, Bernstein BE, Fraenkel E, Cantor AB. 2009. Novel Insights into GATA-1 Mediated Gene Activation versus Repression via Genome-wide Chromatin Occupancy Analysis. Molecular Cell, 36: 682-695. [NIHMS 163436].
Cantor AB, Iwasaki H, Arinobu Y, Moran TB, Shigematsu H, Sullivan MR, Akashi K, Orkin SH. 2008. Antagonism of FOG-1 and GATA factors in fate choice for the mast cell lineage. J. Exp. Med., 205:611-624. PMC ID: PMC 2275384.
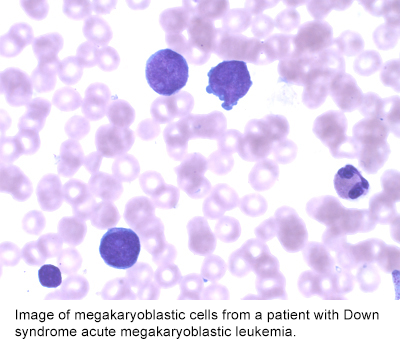
Dysregulated Gene Expression in Human Leukemogenesis
Many of the same transcription factors and associated epigenetic cofactors involved in normal hematopoiesis are mutated in various human leukemias or pre-leukemic conditions. These typically occur as early events in the disease process, providing a “window of opportunity” to intervene in order to reduce the risk of full transformation to leukemia. A major focus of our work in the Cantor Lab is on further understanding the mechanisms by which mutations in these factors contribute to leukemogenesis, and to translate this information into new therapies to prevent leukemia from developing in at-risk individuals.
In particular, we study a number of leukemia predisposition disorders associated with germline and somatic mutations in normal hematopoietic transcription factors, including RUNX1, GATA1, GATA2, and certain ETS factors. Current focus areas of basic/translational research in the Cantor lab on leukemogenesis include:
- understanding the mechanisms by which RUNX1 and GATA2 mutations predispose to clonal hematopoiesis and leukemia development
- identifying small molecules to enhance the residual wild type RUNX1 and GATA2 protein activity in pre-leukemic disorders associated with heterozygous mutations in the RUNX1 and GATA2
- understanding how leukemia-associated mutations in transcriptional regulators alter 3D chromatin structure.
- understanding the mechanisms by which GATA1 mutations lead to Down syndrome Transient Myeloproliferative Disorder and Megakaryoblastic Leukemia.
- understanding molecular mechanisms underlying development of non-MLL rearranged therapy-related myelodysplastic syndrome (t-MDS).
- examining the role of RUNX1 downstream of activated PTPN11/RAS signaling in Juvenile Myelomonocytic Leukemia, and developing RUNX1 inhibitors as potential novel therapeutics for this disease
Woo AJ, Wieland K, Huang H, Akie TE, Piers T, Kim J, Cantor AB. 2013. Developmental Differences in Interferon Signaling Affect GATA1s Induced Megakaryocyte Hyperproliferation, J Clin Invest. 123:3292-3304.
Cantor AB. 2015. Myeloid Proliferations associated with Down Syndrome. Journal of Hematopathology. 8:169-176.
Cantor AB. 2016. Megakaryocytic Transcription Factors in Disease and Leukemia, in Molecular and Cellular Biology of Platelet Formation; ed. Schulze H and Italiano J. Springer International Publishing, Switzerland.
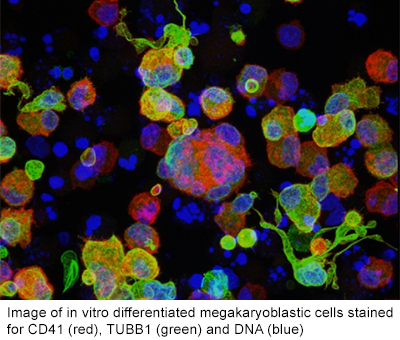
Megakaryopoiesis
The Cantor lab has a special interest in megakaryocytes (Mks), which are the cells that produce blood platelets. Many of the transcription factors that we study play critical roles in Mk differentiation. Compared to other blood lineages, there is much less known about megakaryopoiesis. This is in part due to the rarity of these cells within the bone marrow (~0.5-1% of total nucleated cells) and the relatively recent cloning and purification of the major growth factor for Mks, thrombopoietin. Many outstanding questions remain regarding megakaryopoiesis including understanding the molecular mechanisms governing cell fate choice of bi-potent Mk-erythroid progenitor (MEP) cells for the Mk lineage, the molecular mechanisms that mediate endomitosis (an unusual cell cycle feature in which Mks abort cytokinesis in anaphase B to generate cells of high DNA ploidy), and the mechanisms that regulate proplatelet and platelet formation in vivo. In addition, there are interesting close connections between Mks and hematopoietic stem cells (HSCs) including high overlap of regulatory factors, close developmental hierarchy, and the participation of Mks in the HSC niche. Current focus areas of basic/translational research in the Cantor lab on megakaryopoiesis include:
- identifying the normal in situ signals that stimulate Mk proplatelet formation in vivo.
- enhancing in vitro production of platelet from human induced pluripotent stem cells ultimately for clinical applications.
- further understanding the genetic basis of familial platelet disorders
- understanding the gene regulatory mechanisms that mediate cell fate choice of MEPs for the Mk lineage
Huang H, Yu M, Akie TE, Moran TB, Woo AJ, Tu N, Waldon Z, Lin YY, Steen H, Cantor AB. 2009.Differentiation-dependent Interactions between RUNX-1 and FLI-1 During Megakaryocyte Development, Mol. Cell. Bio. 29:4103-15.
Cantor AB. 2013. Transcriptional Regulation of Megakaryopoiesis, in Atlas on the Cellular and Molecular Development of Human Hematopoiesis, ed. Dame C, Fliedner TM, Sola-Visner M, Yoder M. Springer.
Huang H, Cantor AB. 2009. Common Features of Megakaryocytes and Hematopoietic Stem Cells: What’s the Connection?; J. Cell. Biochem 107:857-64.
Course directed by Dr. Alan Cantor at Harvard Medical School/Division of Medical Sciences
BCMP308qc, Cell Fate Decisions in Development and Disease (Fall semester)
This quarter course offers students an in-depth examination of current knowledge regarding mechanisms of cell fate decisions and cell identity maintenance. In addition, it examines these processes in the context of developmental cell plasticity, cellular reprogramming, and cancer. It is primarily a literature-based course, with examination and discussion of key studies in the field. Concepts involving epigenetics, chromatin remodeling, chromatin accessibility, 3D chromatin architecture, the instructive roles of transcription factors, gene regulatory networks, transcription factor cross-antagonism, feedback loops, multilineage priming, non-coding RNAs, lineage identity maintenance, mitotic bookmarking, lateral inhibition, assymetric division, and niche interactions are explored. These ideas are examined in the context of blood, breast, lung, gastrointestinal tract, and germ cell development.
