Discoveries | Overview
MircoRNA Biogenesis
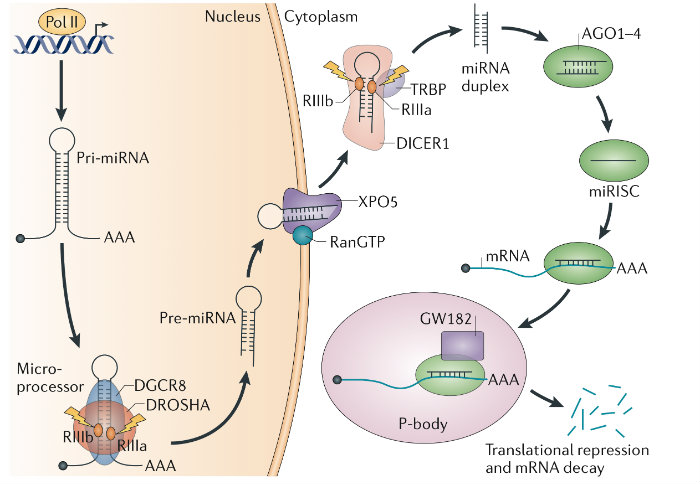
Discovered the Microprocessor and characterized its regulation. For over a decade my work has focused on uncovering the mechanisms of microRNA (miRNA) biogenesis1,2. As a postdoctoral fellow I identified the ‘Microprocessor’ complex that comprises DROSHA and the double-stranded RNA-binding protein DGCR81. DGCR8 maps to a chromosomal region whose monoallelic deletion causes DiGeorge syndrome, the most common human genetic deletion syndrome. Mouse models subsequently revealed that DGCR8 haploinsufficiency contributes to the neuronal and behavioral phenotypes of this disorder. We later uncovered a feedback mechanism for the precise control of DGCR8 expression3. We also discovered that YAP, the downstream target of the tumor-suppressive Hippo-signaling pathway regulates Microprocessor activity in a cell-density dependant manner4. This finding provides a mechanistic explanation for two previously reported phenomena — that cell confluence leads to widespread upregulation of miRNAs, and that tumor cells have a generally lower level of miRNA expression than corresponding normal tissue.
- Gregory R. I.*, Yan K.*, Amuthan G., Chendrimada T., Doratotaj B., Cooch N. and Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004, 432, 235-240. PMID: 15531877 (* equal contribution).
- Gregory R. I., Chendrimada T., Cooch N., and Shiekhattar R. Human RISC couples microRNA maturation and posttranscriptional gene silencing. Cell. 2005, 123, 631-40. PMID: 16271387.
- Triboulet R., Chang H., LaPierre R. J., and Gregory R. I. Posttranscriptional control of DGCR8 expression by the Microprocessor. RNA. 2009, 15, 1005-11. PMID: 19383765. PMC2685516.
- Mori, M., Triboulet, R., Mohseni, M., Schlegelmilc, K., Shrestha, K., Camargo, F. D. Gregory, R. I., Hippo signaling regulates Microprocessor and links cell density-dependent miRNA biogenesis to cancer. Cell 2014. 156, 893-906. PMID: 24581491. PMC3982296.
LIN28/LET-7-Axis
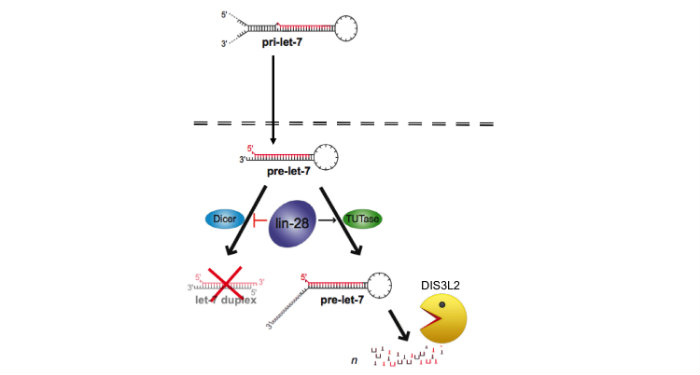
Discovered the LIN28/let-7 axis. We discovered LIN28 as the first negative regulator of the miRNA biogenesis pathway1. Members of the let-7 family are coordinately regulated during development with let-7 expressed at high levels in differentiated cell types. During differentiation of embryonic stem cells (ESCs), mature let-7 miRNAs accumulate but the corresponding primary let-7 transcript remains constant implying posttranscriptional control of miRNA expression, the mechanism for which had remained elusive. We identified the RNA-binding protein LIN28 as a stem cell-specific regulator of miRNA biogenesis. LIN28 was originally discovered as a developmental timing regulator in C. elegans, and later as a reprogramming factor for induced pluripotent stem cells (iPSCs). Since LIN28 mRNA is itself a let-7 target, this LIN28/let-7 axis establishes a double-negative feedback loop, whereby high expression levels of either let-7 or LIN28 will promote a differentiated or embryonic cell fate, respectively. Since the publication of our influential first paper, my laboratory has focused on providing mechanistic insight into this pathway2,3,4. In carrying out these studies, we established a new area of research that profoundly impacts the study of stem cell biology along with many other areas including cancer2. This fundamental discovery launched a new area of research for several other labs, and there is a growing appreciation that the LIN28/let-7 pathway is involved in a sweeping range of biology: germ cell development, somatic cell reprogramming, cancer, aging, inflammation, hematopoiesis, glycolytic metabolism, and diabetes.
- Viswanathan S. R., Daley G. Q., and Gregory R. I. Selective Blockade of MicroRNA Processing by Lin28. Science. 2008, 320, 97-100. PMID: 18292307. PMC3368499.
- Piskounova E., Polytarchou C., Thornton J. E., Hagan J.P., LaPierre R. J., Pothoulakis C., Iliopoulos D., and Gregory R. I. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell. 2011, 147, 1066-79. PMID: 22118463. PMC3227872.
DIS3L2 Mediated Decay
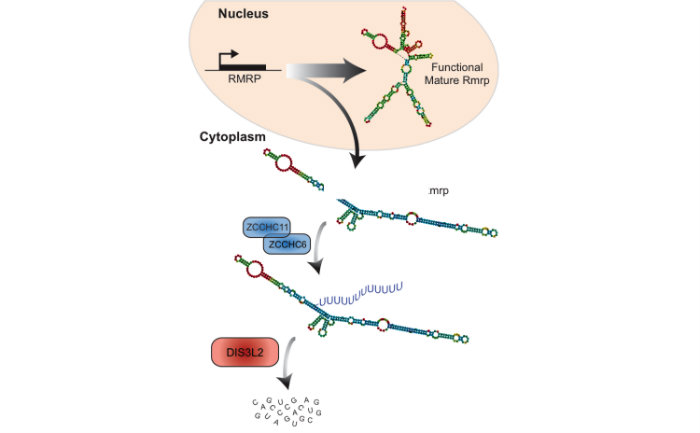
Identified a novel RNA decay pathway in stem cells and cancer. Based on our original discovery that LIN28 selectively controls let-7 expression we sought to better understand the mechanism by which this occurs. This led to our identification that the Terminal Uridylyl Transferase (TUTase), TUT4 (also known as ZCCHC11) is responsible for LIN28-mediated pre-let-7 uridylation and subsequent blockade of let-7 processing1. LIN28-binding to pre-let-7 recruits TUT4 and a related TUTase (TUT7/ZCCHC6) to catalyze pre-let-7 uridylation2. Significantly, this discovery provides the first demonstration that a TUTase can control miRNA biogenesis. We identified DIS3L2 as the 3'-5' exonuclease responsible for the decay of uridylated pre-let-7 in mouse ESCs3. Our paradigm-shifting findings that RNA uridylation triggers degradation by DIS3L2 highlight an important new regulatory mechanism controlling RNA metabolism. Our study also identifies the first physiological RNA substrate of this newly discovered exonuclease, which is mutated in the Perlman syndrome of fetal overgrowth and causes a predisposition to Wilms tumor development. Most recently we applied an unbiased RNA immunoprecipitation strategy to identify Dis3l2 targets in mouse embryonic stem cells. We identified Dis3l2-mediated decay (DMD) as a surveillance pathway for a specific subset of ncRNAs4.
- Hagan J.P., Piskounova E, and Gregory R. I. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nature Structural Molecular Biology. 2009, 16, 1021-5. PMID: 19713958. PMC2758923.
- Thornton J. E., Chang H., Piskounova E., and Gregory R. I. Lin28-mediated control of let-7 microRNA expression by alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7). RNA 2012, 18, 1875-85. PMID: 22898984. PMC3446710.
- Chang H-M., Triboulet R., Thornton J. E,, Gregory R. I. A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature 2013, 497, 244-8. PMID: 23594738, PMC3651781
- Pirouz M., Du P. Munafo M., and Gregory R. I. Dis3l2-Mediated Decay Is a Quality Control Pathway for Noncoding RNAs. Cell Reports 2016, 16, 1861-73. PMID: 27498873. PMC4998061.
- Pirouz M, Ebrahimi A, Gregory RI. Unraveling 3′-end RNA uridylation at nucleotide resolution. Methods 2019. 155 10–19. PMID: 30395968. PMC6387850.
- Mehdi Pirouz, Munafò M, Ebrahimi AG, Choe J, Gregory RI. Exonuclease requirements for mammalian ribosomal RNA biogenesis and surveillance. Nat Struct Mol Biol 2019. 26 490–500. PMID: 31160785. PMC6554070.
-
Pirouz M, Wang CH, Liu Q, Ebrahimi AG, Shamsi F, Tseng YH, Gregory RI. The Perlman syndrome DIS3L2 exoribonuclease safeguards endoplasmic reticulum-targeted mRNA translation and calcium ion homeostasis. Nat Commun 2020. 11 2619. PMID: 32457326. PMC7250864.
Epitranscriptome and Cancer
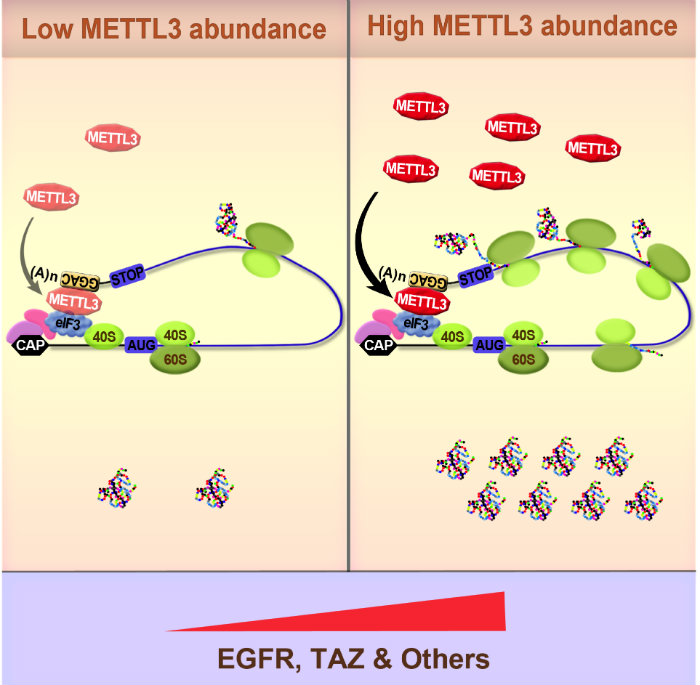
Our most recent work has focused on elucidating the role of RNA methylation in gene expression control. We recently provided the very first functional evidence linking the epitranscriptome with cancer1,2. In these pioneering studies we uncovered a role of the N6-methlyadenosine (m6A) methyltransferase METTL3 in promoting translation of certain oncogenes (including EGFR) to promote lung cancer cell growth, survival, invasion, and tumorigenesis in a mouse xenograft model1,2. Mechanistically we found that METTL3 only promoted translation when bound to the 3’ UTR near a stop codon. Using biochemical approaches we found that METTL3 interacts with the eukaryotic translation initiation factor 3 subunit h (eIF3h) to promote translation of target mRNAs, suggesting an mRNA looping mechanism to facilitate ribosome recycling and translational control. Electron microscopy further supported this model, indicating that METTL3 bound to polyribosomes in close proximity to 5’ cap-binding proteins. METTL3 depletion reduced the translation efficiency of a large subset of mRNAs. We also uncovered the very first m6A epitranscriptomes of human tumor samples2. This provides major insight into the molecular mechanisms and role of m6A in gene regulation and helped establish the epitranscriptome and cancer field.
- Lin, S., Choe, J., Du, P., Triboulet, R. & Gregory, R. I. The m6A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol Cell 2016, 62, 335-345. PMID: 27117702. PMC4860043.
- Choe J., Lin S., Zhang W., Liu Q., Wang L., Ramirez-Moya J., Du P., Kim W., Tang S., Sliz P., Santisteban P., George R. E., Richards W. G., Wong K. K., Locker N., Slack F. J., Gregory R. I. mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature 2018, 561, 556-560.PMID: 30232453. PMC pending.
-
Li J, Gregory R. Mining for METTL3 inhibitors to suppress cancer. Nat Struct Mol Biol 2021, 28 460-462. PMID: 34040230. PMC8197751.
-
Orellana EA, Liu QD, Yankova E, Nelson MD, De Braekeleer E, Zhang W. Kim J, Aspris D, Sendinc E, Garyfallos D, Gu M, Ali R, Gutierrez A, Mikutis S, Bernardes G, Fischer E, Bradley A, Vassiliou A, Slack F, Tzelepis K, Gregory R. METTL1-mediated m7G modification of Arg-TCT tRNA drives oncogenic transformation. Mol Cell 2021. 81 3323-3338. PMID: 34352207. PMC8380730.
-
Ramírez-Moya J, Miliotis C, Baker AR, Gregory RI, Slack FJ, Santisteban P. An ADAR1-dependent RNA editing event in the cyclin-dependent kinase CDK13 promotes thyroid cancer hallmarks. Mol Cancer 2021. 115. PMID: 34496885. PMC8424981.
-
Li J, Gregory R. Mining for METTL3 inhibitors to suppress cancer. Nat Struct Mol Biol 2021, 28 460-462. PMID: 34040230. PMC8197751.
-
Orellana EA, Liu QD, Yankova E, Nelson MD, De Braekeleer E, Zhang W. Kim J, Aspris D, Sendinc E, Garyfallos D, Gu M, Ali R, Gutierrez A, Mikutis S, Bernardes G, Fischer E, Bradley A, Vassiliou A, Slack F, Tzelepis K, Gregory R. METTL1-mediated m7G modification of Arg-TCT tRNA drives oncogenic transformation. Mol Cell 2021. 81 3323-3338. PMID: 34352207. PMC8380730.
-
Ramírez-Moya J, Miliotis C, Baker AR, Gregory RI, Slack FJ, Santisteban P. An ADAR1-dependent RNA editing event in the cyclin-dependent kinase CDK13 promotes thyroid cancer hallmarks. Mol Cancer 2021. 115. PMID: 34496885. PMC8424981.
RNA Methylation
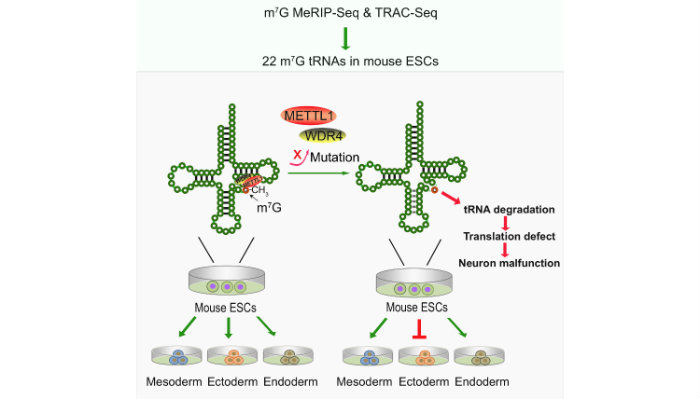
We have developed two independent new methods methods, MeRIP-Seq and TRAC-Seq, to profile the m7G tRNA methylome at single nucleotide resolution. We identified the subset m7G-modified tRNAs and discovered that the METTL1/WDR4-mediated m7G tRNA methylome is required for normal mRNA translation and ESC self-renewal and differentiation3.
- Lin S., Liu Q., Lelyveld V. S., Choe J., Szostak J. W., Gregory R. I. Mettl1/Wdr4-mediated m7G tRNA methylome is required for normal mRNA translation and embryonic stem cell self-renewal and differentiation. Mol Cell 2018, 71, 244-255. PMID: 29983320. PMC6086580.
- Liu Q, Gregory RI. RNAmod: an integrated system for the annotation of mRNA modifications. Nucleic Acids Res 2019. 47(W1):W548-W555. PMID: 31147718. PMC6602476.
- Lin S, Liu Q, Jiang YZ, Gregory RI. Nucleotide resolution profiling of m7G modification by TRAC-Seq. Nat Protoc 2019. 14 3220-3242. PMID: 31619810
- Chen H, Gu L, Orellana EA, Wang Y, Guo J, Liu Q, Wang L, Shen Z, Wu H, Gregory RI, Xing Y, Shi Y. METTL4 is an snRNA m6Am methyltransferase that regulates RNA splicing. Cell Res 2020. 6 544-547. PMID: 31913360 PMC7264358.
- Liu Q, Lang X, Gregory R. An Informatics Pipeline for Profiling and Annotating RNA Modifications. Methods Mol Biol 2021. 2298 15-27. PMID: 34085236.
- Cui J, Liu Q, Sendinc E, Shi Y, Gregory R. Nucleotide resolution profiling of m3C RNA modification by HAC-seq. Nucleic Acids Res 2021. 49: e27. PMID: 33313824. PMC7969016.
- Xiong X, Hou L, Park Y, Molinie B, GTEx Consortium, Gregory R, Kellis M. Genetic drivers of m6A methylation in human brain, lung, heart and muscle. Nature Genet 2021. 53 1156-1165. PMID: 34352207.
- Orellana EA, Siegal E and Gregory RI. tRNA dysregulation and disease. Nature Reviews Genetics. 2022 Nov; 23(11):651-664. PMID: 35681060.
- Yang X, Triboulet R, Liu Q, Sendinc E, Gregory RI.Nat Commun. Exon junction complex shapes the m6A epitranscriptome. 2022 Dec 23;13(1):7904. PMID: 36550132.
- Liu Q, Peng X, Shen M, Qian Q, Xing J, Li C, Gregory RI. Ribo-uORF: a comprehensive data resource of upstream open reading frames (uORFs) based on ribosome profiling. Nucleic Acids Res. 2022 Nov 28 Online ahead of print. PMID: 36440758.
- Li J, Wang L, Hahn Q, Nowak RP, Viennet T, Orellana EA, Roy Burman SS, Yue H, Hunkeler M, Fontana P, Wu H, Arthanari H, Fischer ES, Gregory RI. Structural basis of regulated m7G tRNA modification by METTL1-WDR4. Nature. 2023 Jan 4. Online ahead of print. PMID: 36599985.
MicroRNA Stem Cell Biology
Identified pathways controlling miRNA biogenesis/function and stem cell pluripotency. We identified Trim71 as stem cell specific Ago2-associated protein and found that Trim71 cooperates with miRNAs to repress expression of Cdkn1a, a cyclin-dependent kinase inhibitor that negatively regulates the G1–S transition1. Thus, Trim71 represents a novel ‘stemness’ gene that promotes rapid ESC self-renewal. Our characterization of Ago2 complexes in ESCs led to our discovery that Ago2 protein is unstable and rapidly degraded by the lysosome unless bound by small RNAs and found that the co-chaperones Fkbp4/5 are required for the loading of Ago2 with siRNA and miRNAs2. By exploring the mechanism responsible for the dynamic posttranscriptional control of miR-17~92 during ESC differentiation, we identified a novel miRNA biogenesis intermediate termed progenitor miRNA (pro-miRNA)3. Pri-miR-17~92 adopts an RNA conformation that selectively blocks Microprocessor. Thus, developmentally regulated pro-miRNA processing is key step controlling miRNA expression and explains the posttranscriptional control of miR-17~92 expression in development, thereby revealing a new paradigm for miRNA control. Following up on this dynamic regulation we identified an intermediate ‘poised’ pluripotent state characterized by specific mRNA and miRNA transcriptomes and ability to contribute to mouse chimeras. ISY1-mediated miRNA regulation is necessary and sufficient for establishing poised pluripotency required for naïve-primed transition, providing an explanation for the early embryonic-lethality of global miRNA deficiency4.
- Chang H., Martinez N. J., Thornton J. E., Hagan J. P., Nguyen K. D., and Gregory R. I. Trim71 cooperates with microRNAs to repress Cdkn1a expression and promote embryonic stem cell proliferation. Nature Communications. 2012, 3:923. doi: 10.1038/ncomms1909. PMID: 22735451. PMC3518406.
- Martinez N. J., Chang H-M., Borrajo J., Gregory R. I. The co-chaperones Fkbp4/5 control Argonaute2 expression and facilitate RISC assembly. RNA 2013, 11, 1583-93. PMID: 24049110. PMC3851725.
- Du, P., Wang, L., Sliz, P. & Gregory, R. I. A Biogenesis Step Upstream of Microprocessor Controls miR-17~92 Expression. Cell 2015, 162, 885-899. PMID: 26255770. PMC4537828.
- Du P., Pirouz M., Choi J., Huebner A. J., Clement K., Meissner A., Hochedlinger K., Gregory R. I. An Intermediate Pluripotent State Controlled by MicroRNAs Is Required for the Naive-to-Primed Stem Cell Transition. Cell Stem Cell 2018, 22, 851-64. PMID: 29804889. PMC5990487.
-
Cui Y, Lyu X, Ding L, Ke L, Yang D, Pirouz M, Qi Y, Ong J, Gao G, Du P, Gregory R. Global miRNA dosage control of embryonic germ layer specification. Nature 2021. 593 602-606. PMID: 33953397. PMC8297444.
